Current Projects
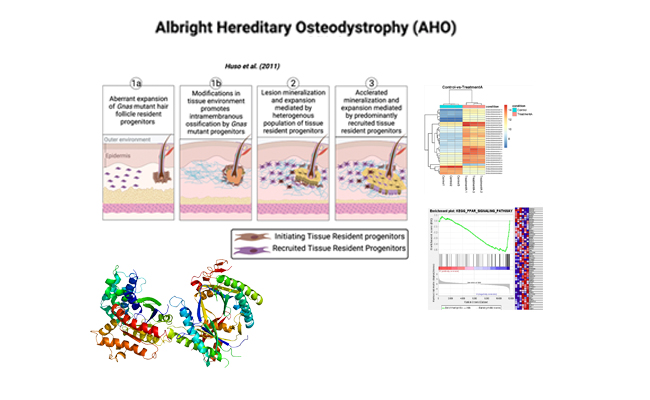
Elucidating the role of GNAS in Albright hereditary osteodystrophy (AHO)
I am currently engaged in a collaborative effort with Dr. Emily Germain-Lee's team, which is dedicated to exploring the function of the GNAS gene that encodes the alpha stimulatory subunit of the G protein in the context of Albright hereditary osteodystrophy (AHO). As part of this project, we have created a mouse model with mutated GNAS and are conducting analyses on biopsied tissues from affected regions. These analyses aim to uncover alterations in gene expression, utilizing RNA sequencing (RNAseq) techniques to gain insights into shifts in gene activity and signaling pathways. .
Tools used: HPC, Cluster, FastQC, Cutadapt, HiSAT2, Stringtie, HTSeq-count, Ballgown, DESeq2, R, Bioconductor

Undestanding the connection between major depressive disorder and Alzheimer’s disease and related dementia (ADRD)
Major depressive disorder (MDD) is one of the most common psychiatric disorders across the lifespan. Besides the disability associated with acute depressive episodes, MDD is also a major risk factor for Alzheimer’s disease and related dementia (ADRD). The mechanisms by which MDD leads to a higher risk of ADRD later in life are poorly understood but probably involve the abnormal regulation of multiple biological processes related to accelerated biological aging (BA). We are investigating the associations of MDD with different biological aging processesand exploring if accelerated BA mediates the association between MDD and the risk of ADRD.
Tools used: R, Bioconductor, IPA, GSEA and others
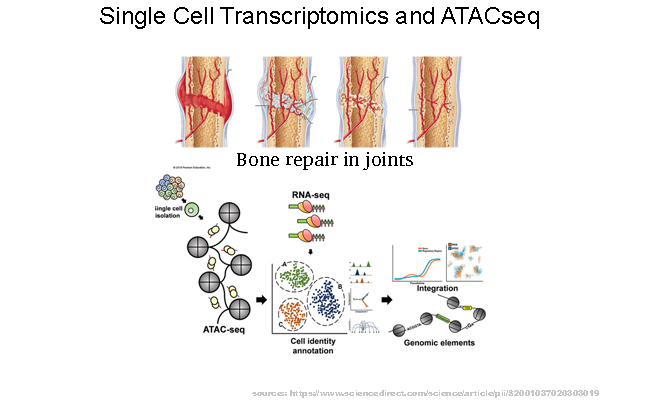
Understanding the process of healing in hip and elbow injury.
In partnership with Professor Guzzo's team at UConn Health, we are delving into the molecular aspects of the healing process after hip and elbow injuries using a mouse model system. Our approach involves employing single-cell multimodal analysis, which combines RNA sequencing (RNAseq) and Assay for Transposase-Accessible Chromatin sequencing (ATAC seq). Through this innovative technique, we aim to uncover intricate changes occurring at the single-cell level, ultimately enhancing our comprehension of the underlying biological mechanisms governing the healing process.
Tools used: CellRanger, R, Bioconductor, Seurat, Monocle, IPA, GSEA and others
Understanding the association between genetic variability within critically ill children and morbidity after PICU discharge.
Stage: Preliminary discussions
In this study we will assess the effects of genetic variation in candidate SNPs on functional status of pediatric survivors of critical illness, focusing on those with acute respiratory failure including children with sepsis and/or pneumonia. Blood plasma profiles will be used in the analysis.
workflow to be finalised.